Getting Bibliographic Metadata¶
As of v0.7, Tethne can parse metadata from the ISI Web of Science, JSTOR’s Data-for-Research portal, and Zotero (RDF). Scopus is not currently supported, but we welcome contributions from those using that database!
Web of Science¶
The ISI Web of Science is a proprietary database owned by Thompson Reuters. If you are affiliated with an academic institution, you may have access to this database via an institutional license.
To access the Web of Science database via the Arizona State University library, find the Web of Science entry in the library’s online catalog. You may be prompted to log in to the University’s Central Authentication System.
Perform a search for literature of interest using the interface provided.
Your search criteria will be informed by the objectives of your research
project. If you are attempting to characterize the development of a research
field, for example, you should choose terms that pick out that field as uniquely
as possible (consider using the Publication Name search field). You can also
pick out literatures originating from particular institutions, by using the
Organization-Enhanced search field.
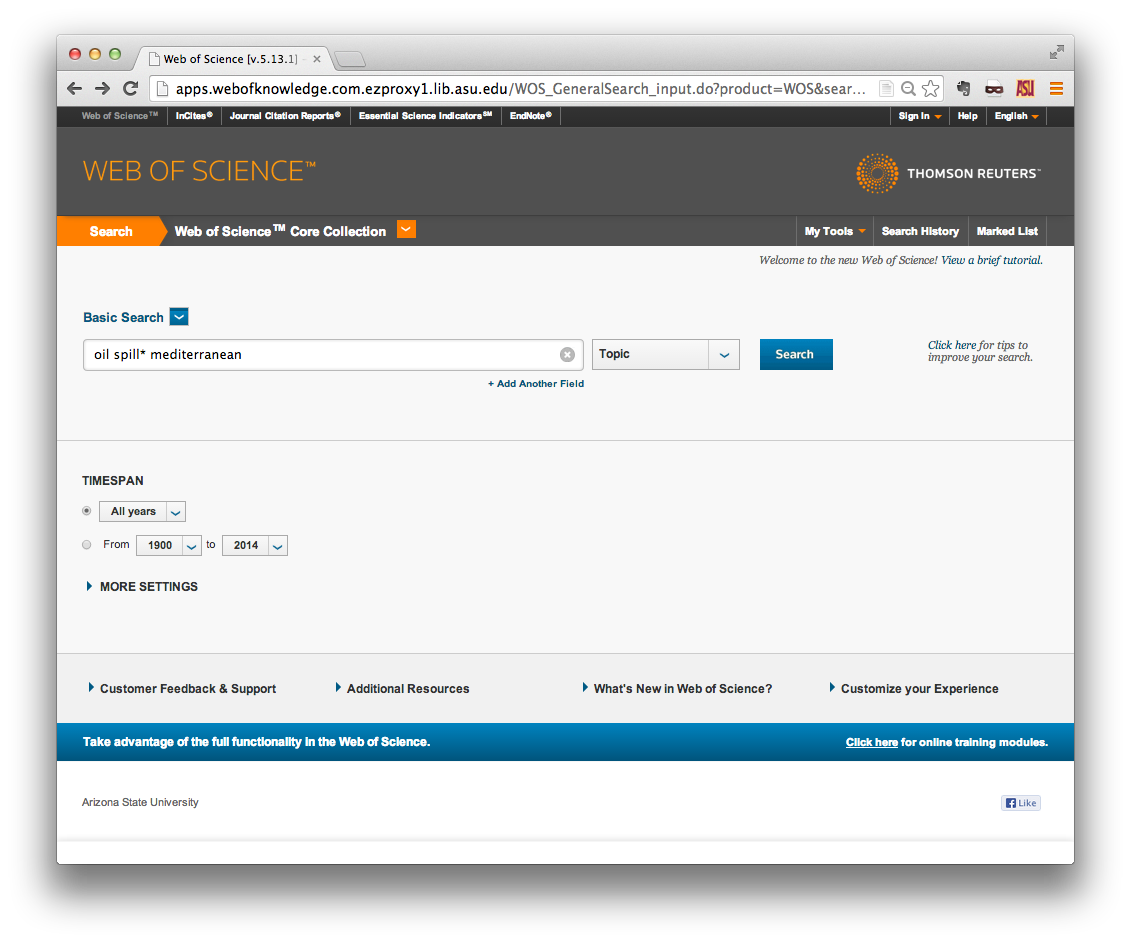
Note also that you can restrict your research to one of three indexes in the Web of Science Core Collection:
- Science Citation Index Expanded is the largest index, containing scientific publications from 1900 onward.
- Social Sciences Citation Index covers 1956 onward.
- Arts & Humanities Citation Index is the smallest index, containing publications from 1975 onward.
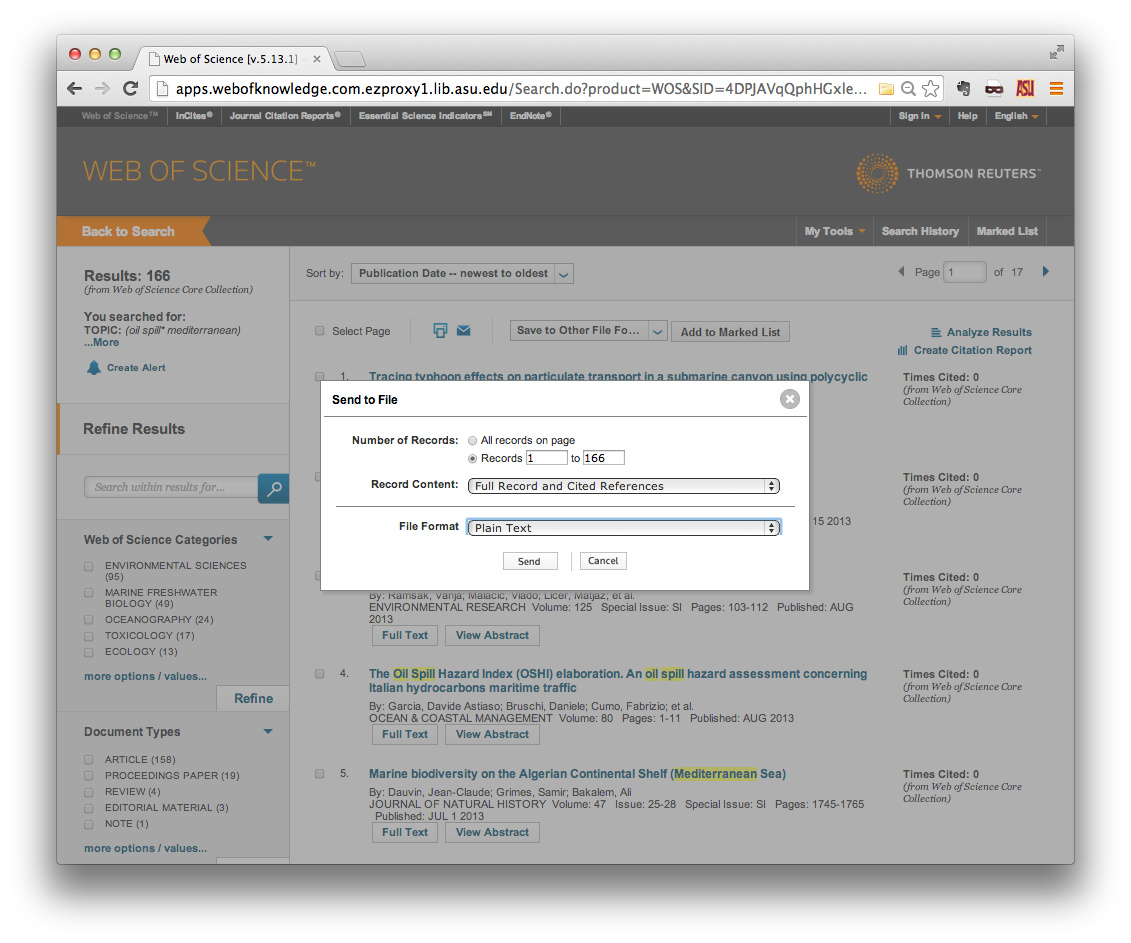
Once you have found the papers that you are interested in, find the Send to:
menu at the top of the list of results. Click the small orange down-arrow, and
select Other File Formats.
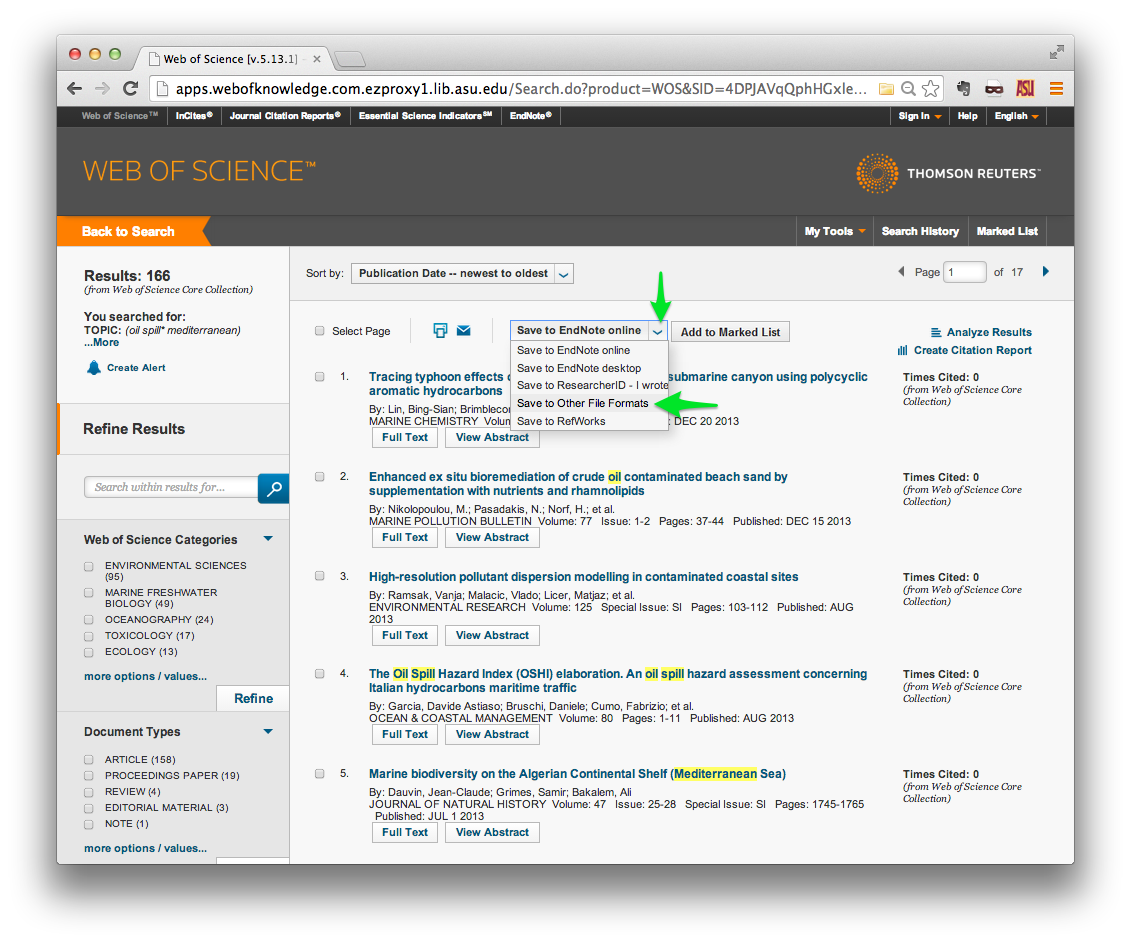
A small in-browser window should open in the foreground. Specify the range of
records that you wish to download. Note that you can only download 500 records
at a time, so you may have to make multiple download requests. Be sure to
specify Full Record and Cited References in the Record Content field, and
Plain Text in the File Format field. Then click Send.
After a few moments, a download should begin. WoS usually returns a field-tagged
data file called savedrecs.txt. Put this in a location on your filesystem
where you can find it later; this is the input for Tethne’s WoS reader methods.
Structure of the WoS Field-Tagged Data File¶
If you open the text file returned by the WoS database (usually named ‘savedrecs.txt’), you should see a whole bunch of field-tagged data. “Field-tagged” means that each metadata field is denoted by a “tag” (a two-letter code), followed by values for that field. A complete list of WoS field tags can be found here. For best results, you should avoid making changes to the contents of WoS data files.
The metadata record for each paper in your data file should begin with:
PT J
...and end with:
ER
There are two author fields: the AU field is always provided, and values take the form “Last, FI”. AF is provided if author full-names are available, and values take the form “Last, First Middle”. For example:
AU Dauvin, JC
Grimes, S
Bakalem, A
AF Dauvin, Jean-Claude
Grimes, Samir
Bakalem, Ali
Citations are listed in the CR block. For example:
CR Airoldi L, 2007, OCEANOGR MAR BIOL, V45, P345
Alexander Vera, 2011, Marine Biodiversity, V41, P545, DOI 10.1007/s12526-011-0084-1
Arvanitidis C, 2002, MAR ECOL PROG SER, V244, P139, DOI 10.3354/meps244139
Bakalem A, 2009, ECOL INDIC, V9, P395, DOI 10.1016/j.ecolind.2008.05.008
Bakalem Ali, 1995, Mesogee, V54, P49
…
Zenetos A, 2005, MEDITERR MAR SCI, V6, P63
Zenetos A, 2004, CIESM ATLAS EXOTIC S, V3
More recent records also include the institutional affiliations of authors in the C1 block.
C1 [Wang, Changlin; Washida, Haruhiko; Crofts, Andrew J.; Hamada, Shigeki;
Katsube-Tanaka, Tomoyuki; Kim, Dongwook; Choi, Sang-Bong; Modi, Mahendra; Singh,
Salvinder; Okita, Thomas W.] Washington State Univ, Inst Biol Chem, Pullman, WA 99164
USA.
For more information about WoS field tags, see a list on the Thompson Reuters website, here.
JSTOR Data-for-Research¶
The JSTOR Data-for-Research (DfR) portal gives researchers access to bibliographic data and N-grams for the entire JSTOR database.
Tethne can use DfR data to generate coauthorship networks, and to improve metadata for Web of Science records. Increasingly, Tethne is also able to use N-gram counts to add information to networks, and can generate corpora for some common topic modeling tools.
Access the DfR portal at http://dfr.jstor.org/ If you don’t already have an account, you will need to create a new account.
After you’ve logged in, perform a search using whatever criteria you please. When you have achieved the result that you desire, create a new dataset request. Under the “Dataset Request” menu in the upper-right corner of the page, click “Submit new request”.
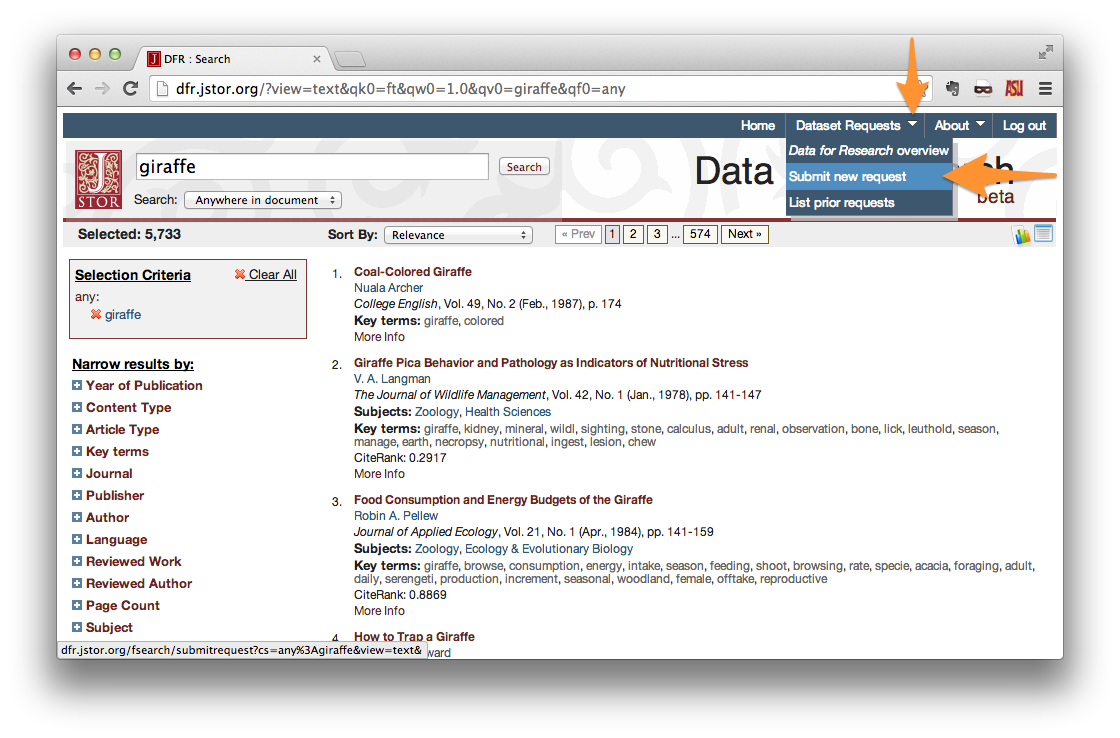
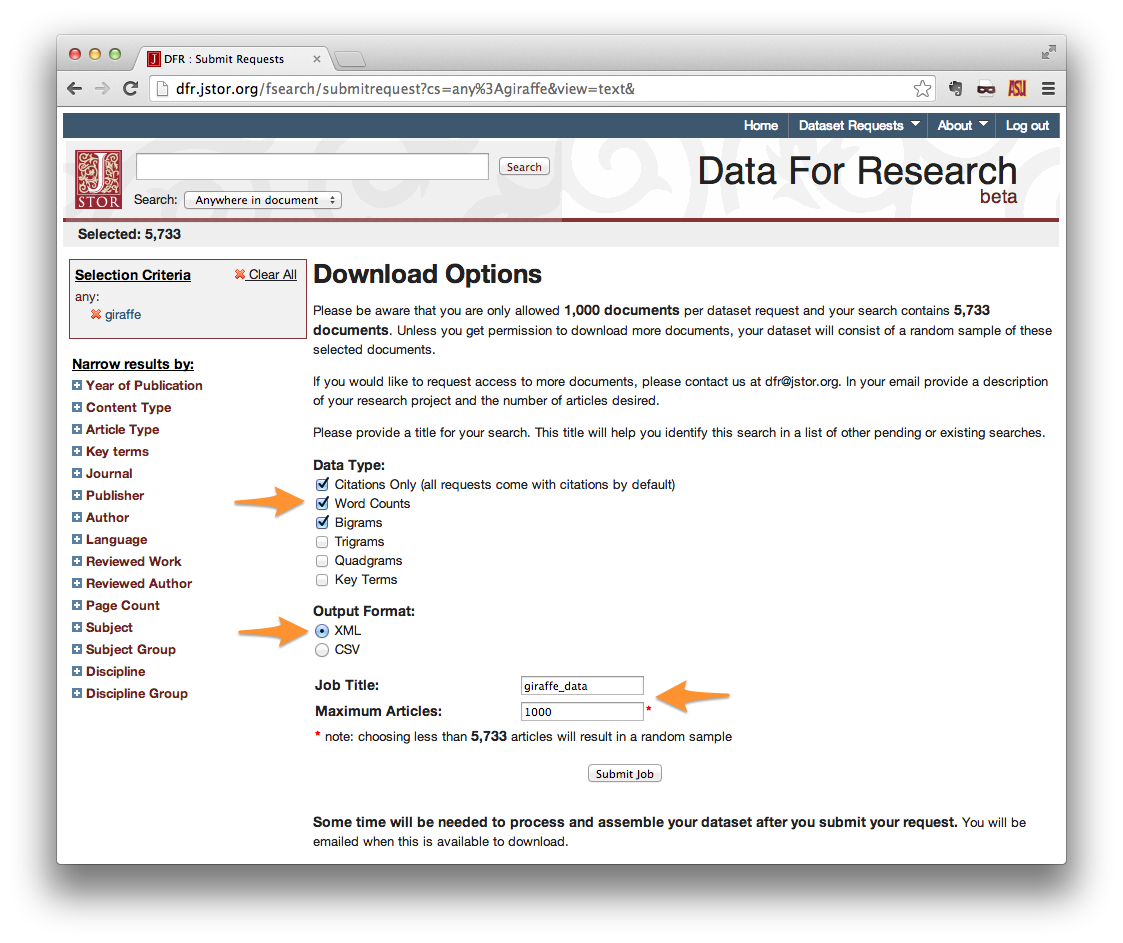
On the Download Options page, select your desired Data Type. If you do not intend to make use of the contents of the papers themselves, then “Citations Only” is sufficient. Otherwise, choose word counts, bigrams, etc.
Output Format should be set to XML.
Give your request a title, and set the maximum number of articles. Note that the maximum documents allowed per request is 1,000. Setting **Maximum Articles* to a value less than the number of search results will yield a random sample of your results.*
Your request should now appear in your list of Data Requests. When your request is ready (hours to days later), you will receive an e-mail with a download link. When downloading from the Data Requests list, be sure to use the link in the full dataset column.
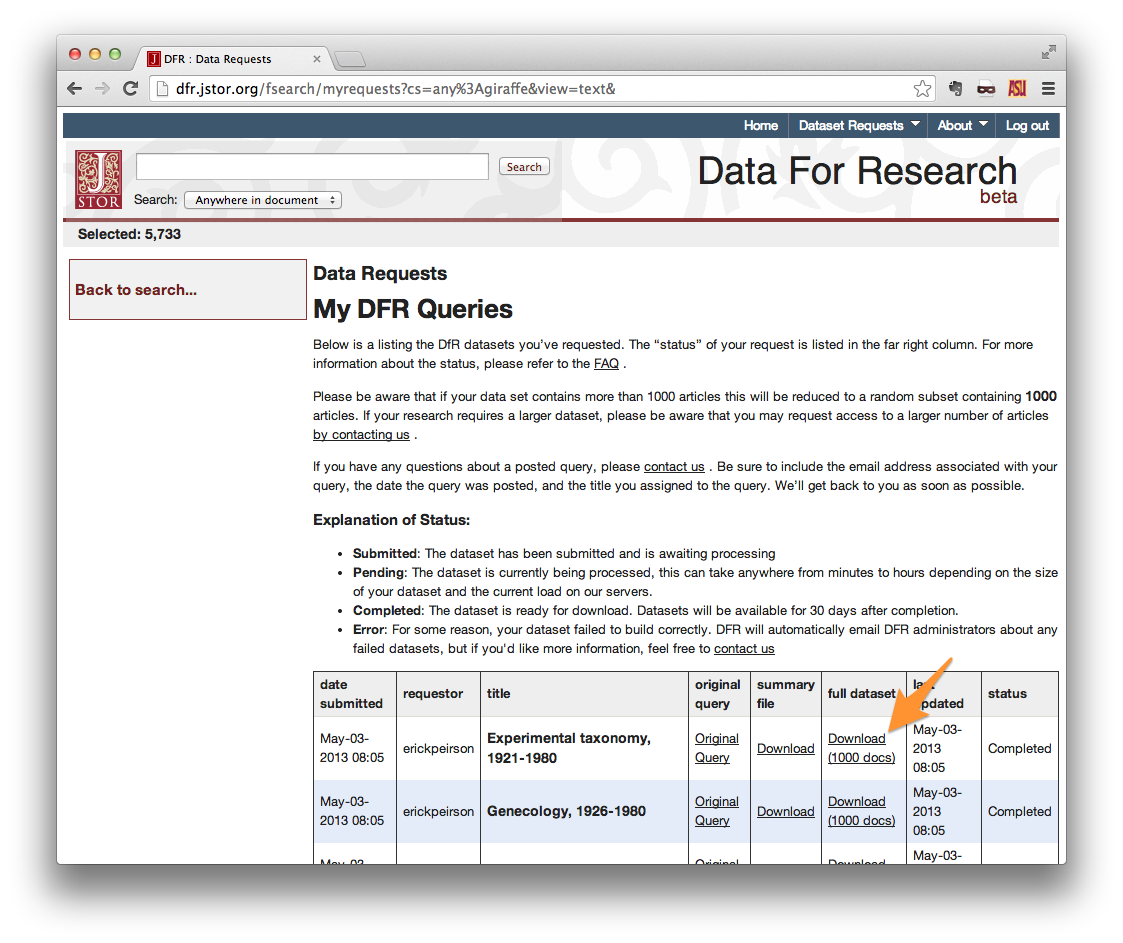
When your dataset download is complete, unzip it. The contents should look something like those shown below.
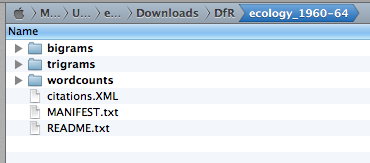
citations.XML contains bibliographic data in XML format. The bigrams,
trigrams, wordcounts folders contain N-gram counts for each document.
In the example above, the path this dataset is
/Users/erickpeirson/Downloads/DfR/ecology_1960-64. This is the path used in
tethne.readers.dfr.read() .
JSTOR Full-Text Data¶
JSTOR occasionally provides full-text data to researchers, upon request. These
data are usually provided in a format similar to the one used for N-grams
(e.g. with a citations.xml metadata file). The text for each document is
stored as a single XML file, the name of which is based on the document’s DOI.
The structure of each document looks something like this:
<?xml version="1.0" encoding="UTF-8"?>
<article id="10.2307/4330485">
<page> <![CDATA[The text for the first page]]> </page>
<page> <![CDATA[The text for the second page]]> </page>
<page> <![CDATA[The text for the third page]]> </page>
</article>
Such a dataset can be parsed just like any DfR dataset, and the document structure (pagination and word order) will be preserved. See ...
Scopus¶
Note
Earlier versions of Tethne had limited support for Scopus. As of 0.8, we’ve moved away from Scopus due to (a) lack of access, and (b) lack of time. If you’re interested in using Scopus data in Tethne, please consider contributing to the project. We’re leaving this documentation here just in case the Scopus reader comes back someday.
Perform your search without whatever parameters you prefer. In this example, we
are searching for documents with phenotypic or plasticity in their
titles, abstracts, or keywords, and we’re searching all available data in
Scopus.
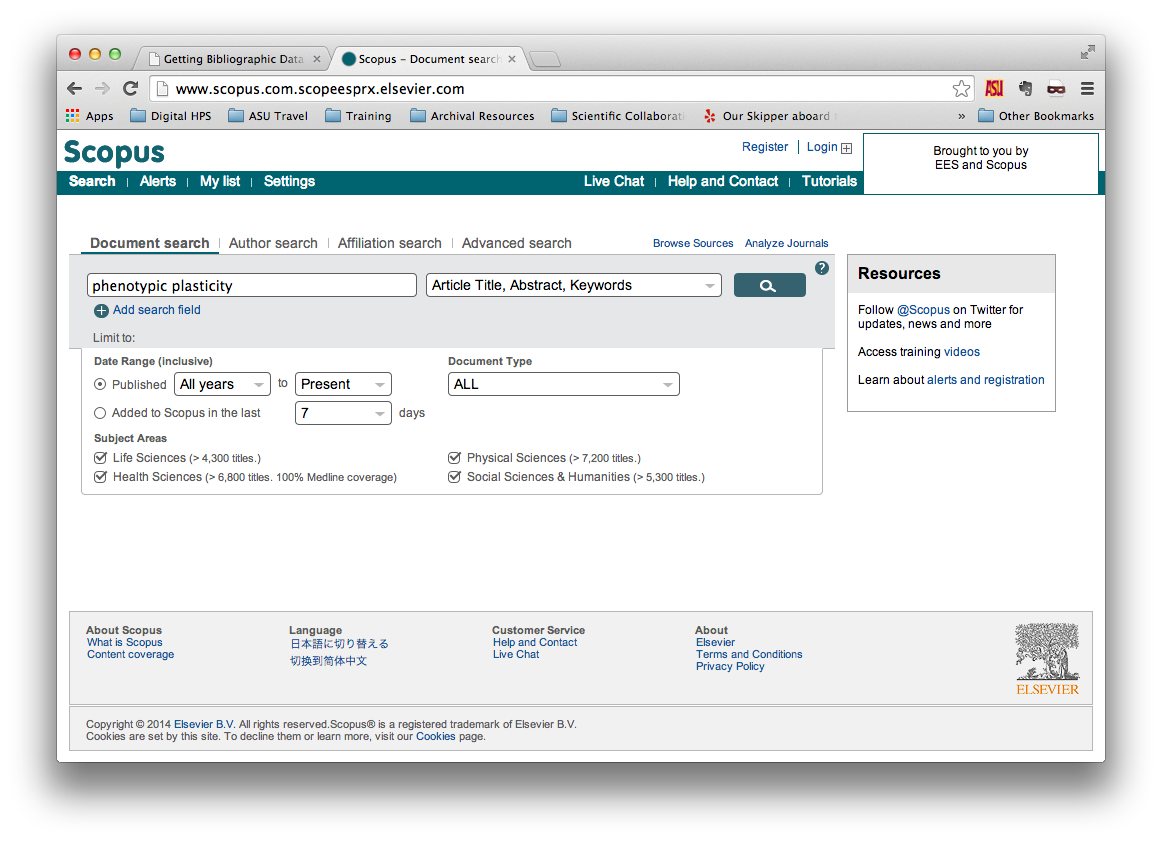
From the search results page, select the records that you wish to export using the checkboxes at left. Then click on “Export”, just above the search results. This should open a menu that looks similar to the one shown below.
Be sure to select the following:
File type: CSV Information to export: All available information
Then click the “Export” button in the bottom right.
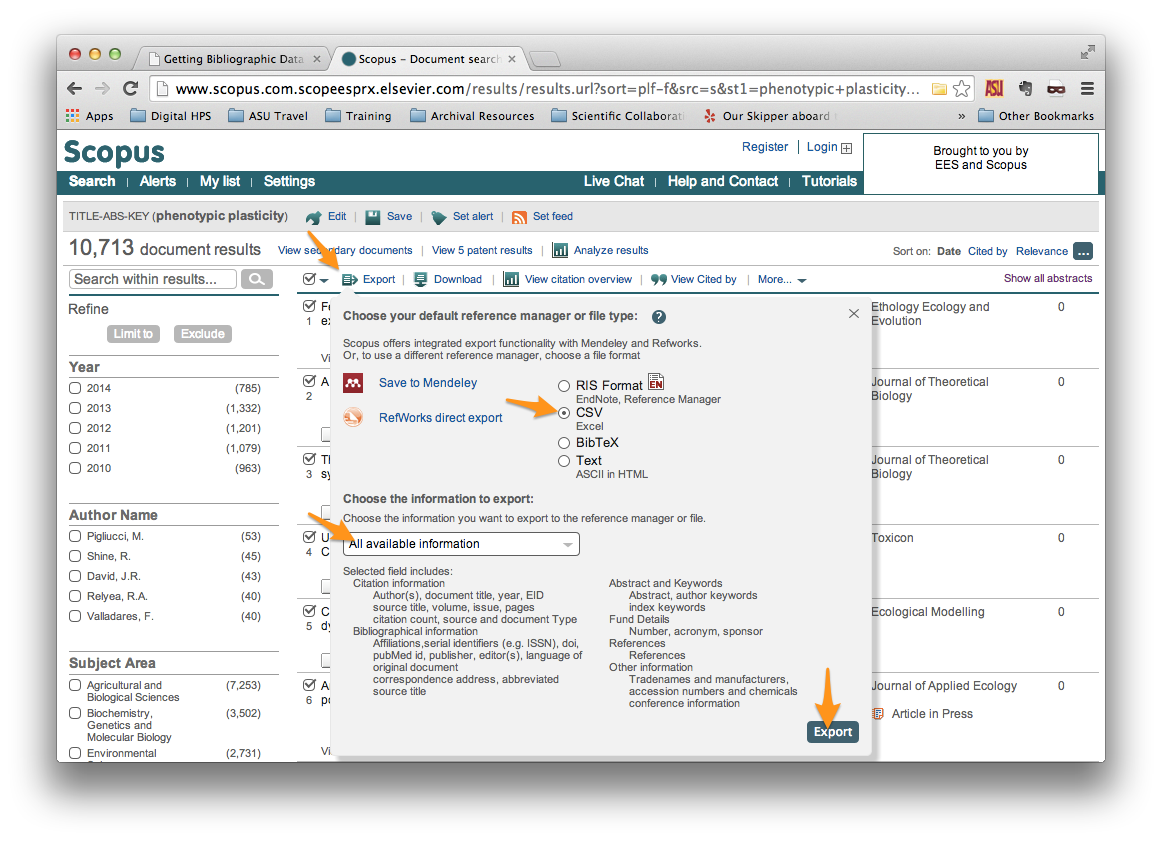
After a few moments, your browser should begin downloading a file called
scopus.csv. If you were to open that file in your favorite spreadsheet
application, the contents should look something like what is shown below.
Zotero¶
Zotero is a tool for collecting and managing bibliographic data. Zotero is especially useful as a data collection tool for computational humanities, since it is quite good at scraping metadata and (when available) full-text documents from databases like Google Scholar, JSTOR, and PubMed.
For an introduction to Zotero, see the Quick Start Guide.
For an introduction to collecting bibliographic metadata and full-text content with Zotero, see this video tutorial. For a text version, see this tutorial.
In order to work with your Zotero collection in Tethne, you will first need to export it in Zotero RDF format.
To export a collection:
- Select the collection from your Library.
- Right-click on the collection, and select “Export Collection”. A small modal dialog should appear.
- Select “Zotero RDF” from the Format field.
- If you want to parse full-text content (e.g. from attached PDF files), check the “Export Files” option.
- Click OK. An Export (save) dialog should appear.
- Navigate to the place on your filesystem to which you wish to export your collection. Put it someplace easy to find, since you’ll need to know the path to your export later on.
- Give your export a name in the “Save As” field.
- Click OK.
This will create a new directory with the name that you specified in step 7.
Inside, you should find a file with the same name, and a .rdf extension. If
you opted to include attached files (step 4), you should also see a folder
called “files”.

