tethne.analyze package¶
Submodules¶
tethne.analyze.collection module¶
Methods for analyzing GraphCollections.
algorithm |
Apply a method from NetworkX to all networkx.Graph objects in the GraphCollection G. |
attachment_probability |
Calculates the observed attachment probability for each node at each time-step. |
connected |
Performs analysis methods from networkx.connected on each graph in the collection. |
delta |
|
node_global_closeness_centrality |
-
tethne.analyze.collection.algorithm(G, method_name, **kwargs)[source]¶ Apply a
methodfrom NetworkX to all networkx.Graph objects in theGraphCollectionG.For options, see the list of algorithms in the NetworkX documentation. Not all of these have been tested.
Parameters: G :
GraphCollectionThe
GraphCollectionto analyze. The specified method will be applied to each graph inG.method_name : string
Name of a method in NetworkX to execute on graph collection.
**kwargs
A list of keyword arguments that should correspond to the parameters of the specified method.
Returns: results : dict
Indexed by element (node or edge) and graph index (e.g.
date).Raises: ValueError
If no such method exists.
Examples
Betweenness centrality: (
Gis aGraphCollection)>>> from tethne.analyze import collection >>> BC = collection.algorithm(G, 'betweenness_centrality') >>> print BC[0] {1999: 0.010101651117889644, 2000: 0.0008689093723107329, 2001: 0.010504898852426189, 2002: 0.009338654511194512, 2003: 0.007519105636349891}
-
tethne.analyze.collection.attachment_probability(G)[source]¶ Calculates the observed attachment probability for each node at each time-step.
Attachment probability is calculated based on the observed new edges in the next time-step. So if a node acquires new edges at time t, this will accrue to the node’s attachment probability at time t-1. Thus at a given time, one can ask whether degree and attachment probability are related.
Parameters: G :
GraphCollectionMust be sliced by ‘date’. See
GraphCollection.slice().Returns: probs : dict
Keyed by index in G.graphs, and then by node.
-
tethne.analyze.collection.connected(G, method_name, **kwargs)[source]¶ Performs analysis methods from networkx.connected on each graph in the collection.
Parameters: G :
GraphCollectionThe
GraphCollectionto analyze. The specified method will be applied to each graph inG.method : string
Name of method in networkx.connected.
**kwargs : kwargs
Keyword arguments, passed directly to method.
Returns: results : dict
Keys are graph indices, values are output of method for that graph.
Raises: ValueError
If name is not in networkx.connected, or if no such method exists.
tethne.analyze.corpus module¶
Methods for analyzing Corpus objects.
burstness |
Estimate burstness profile for the topn features (or flist) in feature. |
feature_burstness |
Estimate burstness profile for a feature over the 'date' axis. |
sigma |
Calculate sigma (from Chen 2009) for all of the nodes in a GraphCollection. |
-
tethne.analyze.corpus.burstness(corpus, featureset_name, features=[], k=5, topn=20, perslice=False, normalize=True, **kwargs)[source]¶ Estimate burstness profile for the
topnfeatures (orflist) infeature.Uses the popular burstness automaton model inroduced by Kleinberg (2002).
Parameters: corpus :
Corpusfeature : str
Name of featureset in
corpus. E.g.'citations'.k : int
(default: 5) Number of burst states.
topn : int or float {0.-1.}
(default: 20) Number (int) or percentage (float) of top-occurring features to return. If
flistis provided, this parameter is ignored.perslice : bool
(default: False) If True, loads
topnfeatures per slice. Otherwise, loadstopnfeatures overall. Ifflistis provided, this parameter is ignored.flist : list
List of features. If provided,
topnandpersliceare ignored.normalize : bool
(default: True) If True, burstness is expressed relative to the hightest possible state (
k-1). Otherwise, states themselves are returned.kwargs : kwargs
Parameters for burstness automaton HMM.
Returns: B : dict
Keys are features, values are tuples of ( dates, burstness )
Examples
>>> from tethne.analyze.corpus import burstness >>> B = burstness(corpus, 'abstractTerms', flist=['process', 'method'] >>> B['process'] ([1990, 1991, 1992, 1993], [0., 0.4, 0.6, 0.])
-
tethne.analyze.corpus.feature_burstness(corpus, featureset_name, feature, k=5, normalize=True, s=1.1, gamma=1.0, **slice_kwargs)[source]¶ Estimate burstness profile for a feature over the
'date'axis.Parameters: corpus :
Corpusfeature : str
Name of featureset in
corpus. E.g.'citations'.findex : int
Index of
featureincorpus.k : int
(default: 5) Number of burst states.
normalize : bool
(default: True) If True, burstness is expressed relative to the hightest possible state (
k-1). Otherwise, states themselves are returned.kwargs : kwargs
Parameters for burstness automaton HMM.
-
tethne.analyze.corpus.sigma(G, corpus, featureset_name, B=None, **kwargs)[source]¶ Calculate sigma (from Chen 2009) for all of the nodes in a
GraphCollection.You can set parameters for burstness estimation using
kwargs:Parameter Description s Scaling parameter ( > 1.)that controls graininess of burst detection. Lower values make the model more sensitive. Defaults to 1.1. gamma Parameter that controls the ‘cost’ of higher burst states. Defaults to 1.0. k Number of burst states. Defaults to 5. Parameters: G :
GraphCollectioncorpus :
Corpusfeature : str
Name of a featureset in corpus.
Examples
Assuming that you have a
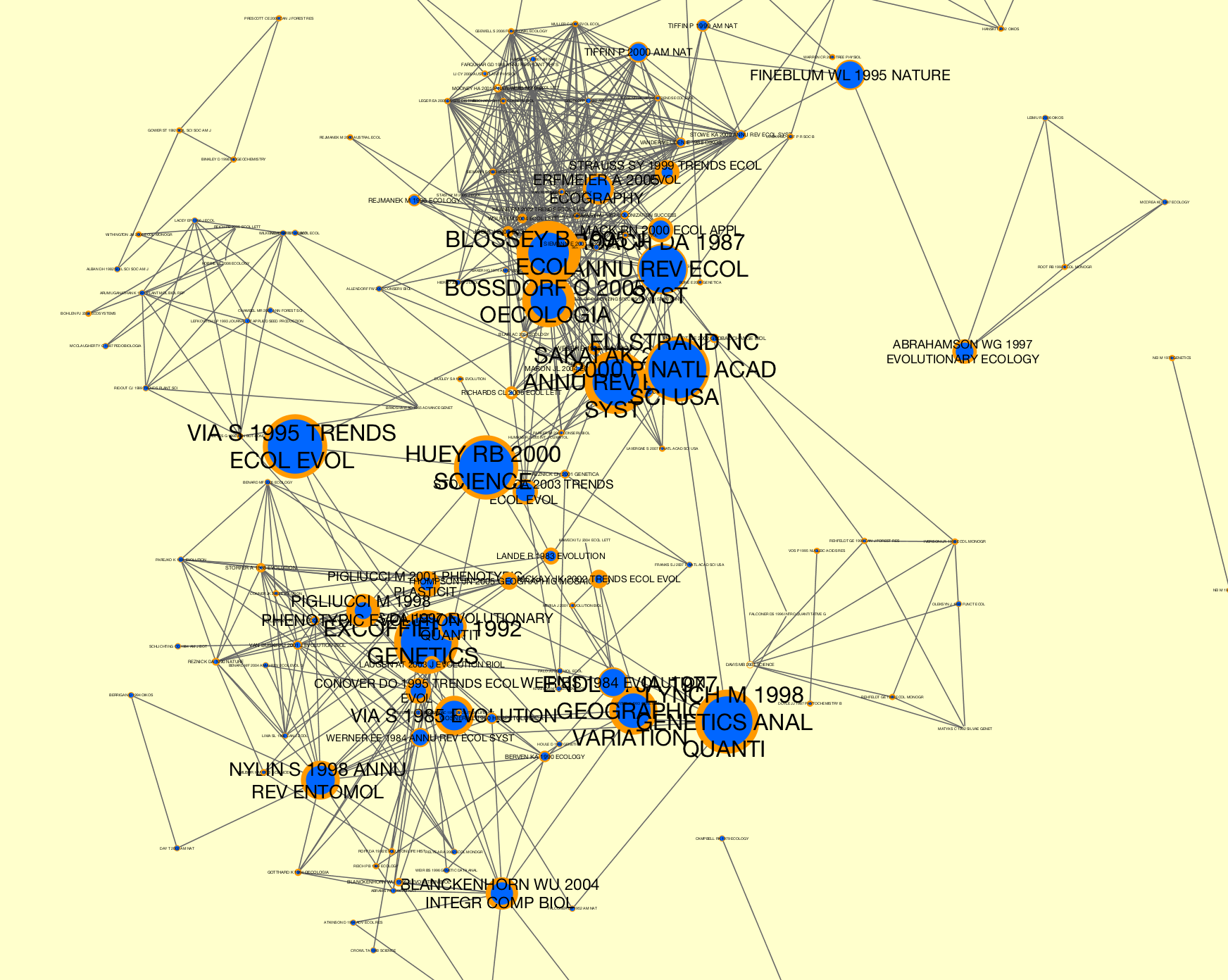
Corpusgenerated from WoS data that has been sliced bydate.>>> # Generate a co-citation graph collection. >>> from tethne import GraphCollection >>> kwargs = { 'threshold':2, 'topn':100 } >>> G = GraphCollection() >>> G.build(corpus, 'date', 'papers', 'cocitation', method_kwargs=kwargs) >>> # Calculate sigma. This may take several minutes, depending on the >>> # size of your co-citaiton graph collection. >>> from tethne.analyze.corpus import sigma >>> G = sigma(G, corpus, 'citations') >>> # Visualize... >>> from tethne.writers import collection >>> collection.to_dxgmml(G, '~/cocitation.xgmml')
In the visualization below, node and label sizes are mapped to
sigma, and border width is mapped tocitations.
tethne.analyze.features module¶
Methods for analyzing featuresets.
cosine_distance |
|
cosine_similarity |
Calculate cosine similarity for sparse feature vectors. |
distance |
|
kl_divergence |
Calculate Kullback-Leibler distance. |
-
tethne.analyze.features.angular_similarity(F_a, F_b)[source]¶ Calculate the angular similarity for sparse feature vectors.
Unlike cosine_similarity, this is a true distance metric.
Parameters: F_a :
FeatureF_b :
FeatureReturns: similarity : float
Cosine similarity.
-
tethne.analyze.features.cosine_similarity(F_a, F_b)[source]¶ Calculate cosine similarity for sparse feature vectors.
Parameters: F_a :
FeatureF_b :
FeatureReturns: similarity : float
Cosine similarity.
tethne.analyze.graph module¶
Methods for network analysis.
global_closeness_centrality |
Calculates global closeness centrality for one or all nodes in the network. |
-
tethne.analyze.graph.global_closeness_centrality(g, node=None, normalize=True)[source]¶ Calculates global closeness centrality for one or all nodes in the network.
See
node_global_closeness_centrality()for more information.Parameters: g : networkx.Graph
normalize : boolean
If True, normalizes centrality based on the average shortest path length. Default is True.
Returns: C : dict
Dictionary of results, with node identifiers as keys and gcc as values.
Module contents¶
Methods for analyzing Corpus, GraphCollection, and
networkx.Graph objects.
collection |
Methods for analyzing GraphCollections. |
corpus |
Methods for analyzing Corpus objects. |
features |
Methods for analyzing featuresets. |
graph |
Methods for network analysis. |