tethne.classes package¶
Submodules¶
tethne.classes.corpus module¶
A Corpus is a container for Papers.
-
class
tethne.classes.corpus.Corpus(papers=[], index_by=None, index_fields=['authors', 'citations', 'ayjid', 'date'], index_features=['authors', 'citations'], **kwargs)[source]¶ Bases:
objectA
Corpusrepresents a collection ofPaperinstances.distributionCalculates the number of papers in each slice, as defined by slice_kwargs.feature_distributionCalculates the distribution of a feature across slices of the corpus. featuresContains FeatureSets for aCorpusinstance.indexIndexes the Papers in thisCorpusinstance by the attributeattr.index_bySpecifies the field in Papers that should be used as the primary indexing field for aCorpusinstance.index_featureCreates a new FeatureSetfrom the attributefeature_namein eachPaper.indexed_papersThe primary index for Papers in aCorpusinstance.indicesContains field indices for the Papers in aCorpusinstance.papersA list of all Papers in theCorpus.selectRetrieves a subset of Papers based on selection criteria.sliceReturns a generator that yields (key, subcorpus)tuples for sequential time windows.subcorpusGenerates a new Corpususing the criteria inselector.top_featuresRetrieves the top topnmost numerous features in the corpus.Corpusobjects are generated by the bibliographic readers in thetethne.readersmodule.>>> from tethne.readers.wos import read >>> read('/path/to/data') <tethne.classes.corpus.Corpus object at 0x10278ea10>
You can also build a
Corpusfrom a list ofPapers.>>> papers = however_you_generate_papers() # <- list of Papers. >>> corpus = Corpus(papers)
All of the
Papers in theCorpuswill be indexed. You can control which field is used for indexing by passing theindex_bykeyword argument to one of thereadmethods or to theCorpusconstructor.>>> corpus = Corpus(papers, index_by='doi') >>> corpus.indexed_papers.keys() ['doi/123', 'doi/456', ..., 'doi/789']
The WoS
readmethod uses thewosidfield by default, and the DfRreadmethod usesdoi. The Zoteroreadmethod tries to use whatever it can find. If the selectedindex_byfield is not set or not available, a unique key will be generated using the title and author names.By default,
Corpuswill also index theauthorsandcitationsfields. To control which fields are indexed, pass theindex_fieldsargument, or callCorpus.index()directly.>>> corpus = Corpus(papers, index_fields=['authors', 'date']) >>> corpus.indices.keys() ['authors', 'date']
Similarly,
Corpuswill index features. By default,authorsandcitationswill be indexed as features (i.e. available for network-building methods). To control which fields are indexed as features, pass theindex_featuresargument, or callCorpus.index_features().>>> corpus = Corpus(papers, index_features=['unigrams']) >>> corpus.features.keys() ['unigrams']
There are a variety of ways to select
Papers from the corpus.>>> corpus = Corpus(papers) >>> corpus[0] # Integer indices yield a single Paper. <tethne.classes.paper.Paper object at 0x103037c10> >>> corpus[range(0,5)] # A list of indices will yield a list of Papers. [<tethne.classes.paper.Paper object at 0x103037c10>, <tethne.classes.paper.Paper object at 0x10301c890>, ... <tethne.classes.paper.Paper object at 0x10302f5d0>] >>> corpus[('date', 1995)] # You can select based on indexed fields. [<tethne.classes.paper.Paper object at 0x103037c10>, <tethne.classes.paper.Paper object at 0x10301c890>, ... <tethne.classes.paper.Paper object at 0x10302f5d0>] >>> corpus['citations', ('DOLE RJ 1952 CELL')] # All papers with this citation! [<tethne.classes.paper.Paper object at 0x103037c10>, <tethne.classes.paper.Paper object at 0x10301c890>, ... <tethne.classes.paper.Paper object at 0x10302f5d0>] >>> corpus[('date', range(1993, 1995))] # Multiple values are supported, too. [<tethne.classes.paper.Paper object at 0x103037c10>, <tethne.classes.paper.Paper object at 0x10301c890>, ... <tethne.classes.paper.Paper object at 0x10302f5d0>]
If you prefer to retrieve a
Corpusrather than simply a list ofPaperinstances (e.g. to build networks), useCorpus.subcorpus().subcorpusaccepts selector arguments just likeCorpus.__getitem__().>>> corpus = Corpus(papers) >>> subcorpus = corpus.subcorpus(('date', 1995)) >>> subcorpus <tethne.classes.corpus.Corpus object at 0x10278ea10>
-
distribution(**slice_kwargs)[source]¶ Calculates the number of papers in each slice, as defined by
slice_kwargs.Parameters: slice_kwargs : kwargs
Keyword arguments to be passed to
Corpus.slice().Returns: list
Examples
>>> corpus.distribution(step_size=1, window_size=1) [5, 5]
-
feature_distribution(featureset_name, feature, mode='counts', **slice_kwargs)[source]¶ Calculates the distribution of a feature across slices of the corpus.
Parameters: featureset_name : str
Name of a
FeatureSetin theCorpus.feature : str
Name of the specific feature of interest. E.g. if
featureset_name='citations', thenfeaturecould be something like'DOLE RJ 1965 CELL'.mode : str
(default:
'counts') If set to'counts', values will be the sum of all count values for the feature in each slice. If set to'documentCounts', values will be the number of papers in which the feature occurs in each slice.slice_kwargs : kwargs
Keyword arguments to be passed to
Corpus.slice().Returns: list
Examples
>>> corpus.feature_distribution(featureset_name='citations', ... feature='DOLE RJ 1965 CELL', ... step_size=1, window_size=1) [2, 15, 25, 1]
-
features= {}¶ Contains
FeatureSets for aCorpusinstance.New
FeatureSets can be created from attributes ofPaperusingindex_feature().
-
index(attr)[source]¶ Indexes the
Papers in thisCorpusinstance by the attributeattr.New indices are added to
indices.Parameters: attr : str
The name of a
Paperattribute.
-
index_by= None¶ Specifies the field in
Papers that should be used as the primary indexing field for aCorpusinstance.
-
index_class¶ alias of
dict
-
index_feature(feature_name, tokenize=<function <lambda>>, structured=False)[source]¶ Creates a new
FeatureSetfrom the attributefeature_namein eachPaper.New
FeatureSets are added tofeatures.Parameters: feature_name : str
The name of a
Paperattribute.
-
index_kwargs= {}¶
-
index_paper_by_feature(paper, feature_name, tokenize=<function <lambda>>, structured=False)[source]¶
-
indexed_papers= {}¶ The primary index for
Papers in aCorpusinstance. Keys are based onindex_by, and values arePaperinstances.
-
indices= {}¶ Contains field indices for the
Papers in aCorpusinstance.The
'citations'index, for example, allows us to look up all of the Papers that contain a particular bibliographic reference:>>> for citation, papers in corpus.indices['citations'].items()[7:10]: ... print 'The following Papers cite {0} \n\n\t{1} \n'.format(citation, '\n\t'.join(papers)) The following Papers cite WHITFIELD J 2006 NATURE: WOS:000252758800011 WOS:000253464000004 The following Papers cite WANG T 2006 GLOBAL CHANGE BIOL: WOS:000282225000019 WOS:000281546800001 WOS:000251903200006 WOS:000292901400010 WOS:000288656800015 WOS:000318353300001 WOS:000296710600017 WOS:000255552100006 WOS:000272153800012 The following Papers cite LINKOSALO T 2009 AGR FOREST METEOROL: WOS:000298398700003
Notice that the values above are not Papers themselves, but identifiers. These are the same identifiers used in the primary index, so we can use them to look up
Papers:>>> papers = corpus.indices['citations']['CARLSON SM 2004 EVOL ECOL RES'] # Who cited Carlson 2004? >>> print papers >>> for paper in papers: ... print corpus.indexed_papers[paper] ['WOS:000311994600006', 'WOS:000304903100014', 'WOS:000248812000005'] <tethne.classes.paper.Paper object at 0x112d1fe10> <tethne.classes.paper.Paper object at 0x1121e8310> <tethne.classes.paper.Paper object at 0x1144ad390>
You can create new indices using
index().
-
select(selector, index_only=False)[source]¶ Retrieves a subset of
Papers based on selection criteria.There are a variety of ways to select
Papers.>>> corpus = Corpus(papers) >>> corpus[0] # Integer indices yield a single Paper. <tethne.classes.paper.Paper object at 0x103037c10> >>> corpus[range(0,5)] # A list of indices yields a list of Papers. [<tethne.classes.paper.Paper object at 0x103037c10>, <tethne.classes.paper.Paper object at 0x10301c890>, ... <tethne.classes.paper.Paper object at 0x10302f5d0>] >>> corpus[('date', 1995)] # Select based on indexed fields. [<tethne.classes.paper.Paper object at 0x103037c10>, <tethne.classes.paper.Paper object at 0x10301c890>, ... <tethne.classes.paper.Paper object at 0x10302f5d0>] >>> corpus['citations', ('DOLE RJ 1952 CELL')] # Citing papers! [<tethne.classes.paper.Paper object at 0x103037c10>, <tethne.classes.paper.Paper object at 0x10301c890>, ... <tethne.classes.paper.Paper object at 0x10302f5d0>] >>> corpus[('date', range(1993, 1995))] # Multiple values are OK. [<tethne.classes.paper.Paper object at 0x103037c10>, <tethne.classes.paper.Paper object at 0x10301c890>, ... <tethne.classes.paper.Paper object at 0x10302f5d0>]
If you prefer to retrieve a
Corpusrather than simply a list ofPaperinstances (e.g. to build networks), useCorpus.subcorpus().Parameters: selector : object
See method description.
Returns: list
A list of
Papers.
-
slice(window_size=1, step_size=1, cumulative=False, count_only=False, subcorpus=True, feature_name=None)[source]¶ Returns a generator that yields
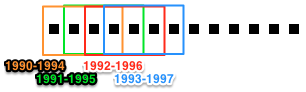
(key, subcorpus)tuples for sequential time windows.Two common slicing patterns are the “sliding time-window” and the “time-period” patterns. Whereas time-period slicing divides the corpus into subcorpora by sequential non-overlapping time periods, subcorpora generated by time-window slicing can overlap.
Sliding time-window – Set
step_size=1, andwindow_sizeto the desired value. Time-period –step_sizeandwindow_sizeshould have the same value.The value of
keyis always the first year in the slice.Parameters: window_size : int
(default: 1) Size of the time window, in years.
step_size : int
(default: 1) Number of years to advance window at each step.
Returns: generator
Examples
>>> from tethne.readers.wos import read >>> corpus = read('/path/to/data') >>> for key, subcorpus in corpus.slice(): ... print key, len(subcorpus) 2005, 5 2006, 5
-
subcorpus(selector)[source]¶ Generates a new
Corpususing the criteria inselector.Accepts selector arguments just like
Corpus.select().>>> corpus = Corpus(papers) >>> subcorpus = corpus.subcorpus(('date', 1995)) >>> subcorpus <tethne.classes.corpus.Corpus object at 0x10278ea10>
-
top_features(featureset_name, topn=20, by='counts', perslice=False, slice_kwargs={})[source]¶ Retrieves the top
topnmost numerous features in the corpus.Parameters: featureset_name : str
Name of a
FeatureSetin theCorpus.topn : int
(default:
20) Number of features to return.by : str
(default:
'counts') If'counts', uses the sum of feature count values to rank features. If'documentCounts', uses the number of papers in which features occur.perslice : bool
(default: False) If True, retrieves the top
topnfeatures in each slice.slice_kwargs : kwargs
If
perslice=True, these keyword arguments are passed toCorpus.slice().
-
tethne.classes.feature module¶
Classes in this module provide structures for additional data about
Papers.
-
class
tethne.classes.feature.BaseFeatureSet(features={})[source]¶ Bases:
object-
N_documents¶
-
N_features¶
-
top(topn, by='counts')[source]¶ Get the top
topnfeatures in theFeatureSet.Parameters: topn : int
Number of features to return.
by : str
(default: ‘counts’) How features should be sorted. Must be ‘counts’ or ‘documentcounts’.
Returns: list
-
unique¶ The set of unique elements in this
FeatureSet.
-
-
class
tethne.classes.feature.Feature(data)[source]¶ Bases:
listA
Featureinstance is a sparse vector of features over a given concept (usually aPaper).For example, a
Featuremight represent word counts for a singlePaper.A
Featuremay be initialized from a list of(feature, value)tuples...>>> myFeature = Feature([('the', 2), ('pine', 1), ('trapezoid', 5)])
...or by passing a list of raw feature tokens:
To get the set of unique features in this
Feature, use :prop:`.Feature.unique`:>>> myFeature.unique set(['the', 'pine', 'trapezoid'])
Normalized feature values (so that all values sum to 1.) can be accessed using :prop:`.Feature.norm`.
>>> myFeature.norm [('the', 0.25), ('pine', 0.125), ('trapezoid', 0.625)]
-
norm¶
-
-
class
tethne.classes.feature.FeatureSet(features=None)[source]¶ Bases:
tethne.classes.feature.BaseFeatureSetA
FeatureSetorganizes multipleFeatureinstances.-
to_gensim_corpus(raw=False)[source]¶ Yield a bag-of-words corpus compatible with the Gensim package.
Returns a (corpus, index) tuple (see below).
Parameters: context : str
If provided, each “document” in the Gensim corpus will be a chunk of type
context.raw : bool
(default: False) If True, documents will be sequences of tokens rather than sequences of (term, count) “bag-of-words” tuples.
Returns: list
A list of lists of (id, count) tuples. Each sub-list represents a single context item (e.g. a document, or a paragraph). This is the “bag of words” representation used in Gensim.
dict
Maps integer IDs to words.
Examples
>>> from tethne.readers.wos import read >>> corpus = read('/path/to/my/data') >>> from nltk.tokenize import word_tokenize >>> corpus.index_feature('abstract', word_tokenize) >>> gensim_corpus, id2word = corpus.features['abstract'].to_gensim_corpus() >>> from gensim import corpora, models >>> model = models.ldamodel.LdaModel(corpus=gensim_corpus, id2word=id2word, num_topics=5, update_every=1, chunksize=100, passes=1)
-
transform(func)[source]¶ Apply a transformation to tokens in this
FeatureSet.Parameters: func : callable
Should take four parameters: token, value in document (e.g. count), value in
FeatureSet(e.g. overall count), and document count (i.e. number of documents in which the token occurs). Should return a new numeric (int or float) value, or None. If value is 0 or None, the token will be excluded.Returns: Examples
Apply a tf*idf transformation.
>>> words = corpus.features['words'] >>> def tfidf(f, c, C, DC): ... tf = float(c) ... idf = log(float(len(words.features))/float(DC)) ... return tf*idf >>> corpus.features['words_tfidf'] = words.transform(tfidf)
-
-
class
tethne.classes.feature.StructuredFeature(tokens, contexts=None, reference=None)[source]¶ Bases:
listA
StructuredFeaturerepresents the contents of a document as an array of tokens, divisible into a set of nested contexts.The canonical use-case is to represent a document as a set of words divided into sentences, paragraphs, and (perhaps) pages.
Parameters: tokens : list
An ordered list of tokens.
contexts : list
A list of (name, indices) 2-tuples, where
nameis string-like and indices is an iterable of int token indices.reference : tuple
A (feature, map) 2-tuple, where
featureis aStructuredFeatureandmapis a dict mapping token indices in thisStructuredFeatureto token indices infeature.-
add_context(name, indices, level=None)[source]¶ Add a new context level to the hierarchy.
By default, new contexts are added to the lowest level of the hierarchy. To insert the context elsewhere in the hierarchy, use the
levelargument. For example,level=0would insert the context at the highest level of the hierarchy.Parameters: name : str
indices : list
Token indices at which each chunk in the context begins.
level : int
Level in the hierarchy at which to insert the context. By default, inserts context at the lowest level of the hierarchy
-
context_chunk(context, j)[source]¶ Retrieve the tokens in the
j``th chunk of context ``context.Parameters: context : str
Context name.
j : int
Index of a context chunk.
Returns: chunk : list
List of tokens in the selected chunk.
-
-
class
tethne.classes.feature.StructuredFeatureSet(features={})[source]¶ Bases:
tethne.classes.feature.BaseFeatureSetA
StructuredFeatureSetorganizes severalStructuredFeatureinstances.-
context_chunks(context=None)[source]¶ Retrieves all tokens, divided into the chunks in context
context.If
contextis not found in a feature, then the feature will be treated as a single chunk.Parameters: context : str
Context name.
Returns: papers : list
2-tuples of (paper ID, chunk indices).
chunks : list
Each item in
chunksis a list of tokens.
-
to_gensim_corpus(context=None, raw=False)[source]¶ Yield a bag-of-words corpus compatible with the Gensim package.
Returns a (corpus, index) tuple (see below).
Parameters: context : str
If provided, each “document” in the Gensim corpus will be a chunk of type
context.raw : bool
(default: False) If True, documents will be sequences of tokens rather than sequences of (term, count) “bag-of-words” tuples.
Returns: list
A list of lists of (id, count) tuples. Each sub-list represents a single context item (e.g. a document, or a paragraph). This is the “bag of words” representation used in Gensim.
dict
Maps integer IDs to words.
Examples
>>> from tethne.readers.wos import read >>> corpus = read('/path/to/my/data') >>> from nltk.tokenize import word_tokenize >>> corpus.index_feature('abstract', word_tokenize, structured=True) >>> gensim_corpus, id2word = corpus.features['abstract'].to_gensim_corpus() >>> from gensim import corpora, models >>> model = models.ldamodel.LdaModel(corpus=gensim_corpus, id2word=id2word, num_topics=5, update_every=1, chunksize=100, passes=1)
-
-
tethne.classes.feature.argsort(l)¶
tethne.classes.graphcollection module¶
A GraphCollection is a set of graphs generated from a
Corpus or model.
-
class
tethne.classes.graphcollection.GraphCollection(corpus=None, method=None, slice_kwargs={}, method_kwargs={}, directed=False)[source]¶ Bases:
dictA
GraphCollectionis an indexed set of networkx.Graphs.When you add a networkx.Graph, the nodes are indexed and relabeled.
>>> from tethne import GraphCollection >>> import networkx as nx >>> G = GraphCollection() >>> g = nx.Graph() >>> g.add_node('A', yes='no') >>> g.add_edge('A', 'B', c='d') >>> G['graph1'] = g # You can also use G.add('graph1', g) >>> G.graph1.nodes(data=True) [(0, {}), (1, {'yes': 'no'})] >>> G.node_index, G.node_lookup ({0: 'B', 1: 'A', -1: None}, {'A': 1, None: -1, 'B': 0})
To build a
GraphCollectionfrom aCorpus, pass it and a method to the constructor, or useGraphCollection.build().>>> corpus = read(datapath) >>> G = GraphCollection(corpus, coauthors) >>> G.build(corpus, authors)
-
add(name, graph)[source]¶ Index and add a networkx.Graph to the
GraphCollection.Parameters: name : hashable
Unique name used to identify the graph.
graph : networkx.Graph
Raises: ValueError
If name has already been used in this
GraphCollection.
-
analyze(method_name, mapper=<built-in function map>, invert=False, **kwargs)[source]¶ Apply a method from NetworkX to each of the graphs in the
GraphCollection.Parameters: method : str or iterable
Must be the name of a method accessible directly from the networkx namespace. If an iterable, should be the complete dot-path to the method, e.g.
nx.connected.is_connectedwould be written as['connected', 'is_connected'].mapper : func
A mapping function. Be default uses Python’s builtin
mapfunction. MUST return results in order.results_by : str
(default: ‘graph’). By default, the top-level key in the results are graph names. If results_by=’node’, node labels are used as top-level keys.
kwargs : kwargs
Any additional kwargs are passed to the NetworkX method.
Returns: dict
Examples
>>> G.analyze('betweenness_centrality') {'test': {0: 1.0, 1: 0.0, 2: 0.0}, 'test2': {0: 0.0, 1: 0.0, 2: 0.0, 3: 0.0}} >>> G.analyze('betweenness_centrality', results_by='node') {0: {'test': 1.0, 'test2': 0.0}, 1: {'test': 0.0, 'test2': 0.0}, 2: {'test': 0.0, 'test2': 0.0}, 3: {'test2': 0.0}}
-
build(corpus, method, slice_kwargs={}, method_kwargs={})[source]¶ Generate a set of networkx.Graphs using
methodon the slices incorpus.Parameters: corpus :
Corpusmethod : str or func
If str, looks for
methodin thetethnenamespace.slice_kwargs : dict
Keyword arguments to pass to
corpus‘slicemethod.method_kwargs : dict
Keyword arguments to pass to
methodalong withcorpus.
-
collapse(weight_attr='weight')[source]¶ Returns a networkx.Graph or
networkx.DiGraphin which the edges between each pair of nodes are collapsed into a single weighted edge.
-
edge_history(source, target, attribute)[source]¶ Returns a dictionary of attribute vales for each Graph in the
GraphCollectionfor a single edge.Parameters: source : str
Identifier for source node.
target : str
Identifier for target node.
attribute : str
The attribute of interest; e.g. ‘betweenness_centrality’
Returns: history : dict
-
edges(data=False, native=True)[source]¶ Returns a list of all edges in the
GraphCollection.Parameters: data : bool
(default: False) If True, returns a list of 3-tuples containing source and target node labels, and attributes.
Returns: edges : list
-
index(name, graph)[source]¶ Index any new nodes in graph, and relabel the nodes in graph using the index.
Parameters: name : hashable
Unique name used to identify the graph.
graph : networkx.Graph
Returns: indexed_graph : networkx.Graph
-
node_history(node, attribute)[source]¶ Returns a dictionary of attribute values for each networkx.Graph in the
GraphCollectionfor a single node.Parameters: node : str
The node of interest.
attribute : str
The attribute of interest; e.g. ‘betweenness_centrality’
Returns: history : dict
-
nodes(data=False, native=True)[source]¶ Returns a list of all nodes in the
GraphCollection.Parameters: data : bool
(default: False) If True, returns a list of 2-tuples containing node labels and attributes.
Returns: nodes : list
-
order(piecewise=False)[source]¶ Returns the total number of nodes in the
GraphCollection.
-
size(piecewise=False)[source]¶ Returns the total number of edges in the
GraphCollection.
-
union(weight_attr='_weight')[source]¶ Returns the union of all graphs in this
GraphCollection.The number of graphs in which an edge exists between each node pair u and v is stored in the edge attribute given be weight_attr (default: _weight).
Parameters: weight_attr : str
(default: ‘_weight’) Name of the edge attribute used to store the number of graphs in which an edge exists between node pairs.
Returns: graph : networkx.Graph
-
tethne.classes.paper module¶
A Paper represents a single bibliographic record.
-
class
tethne.classes.paper.Paper[source]¶ Bases:
objectTethne’s representation of a bibliographic record.
Fields can be set using dict-like assignment, and accessed as attributes.
>>> myPaper = Paper() >>> myPaper['date'] = 1965 >>> myPaper.date 1965
-
ayjid¶ Fuzzy WoS-style identifier, comprised of first author’s name (LAST I), pubdate, and journal.
Returns: ayjid : str
-
citations¶
-
tethne.classes.streaming module¶
-
class
tethne.classes.streaming.StreamingCorpus(*args, **kwargs)[source]¶ Bases:
tethne.classes.corpus.CorpusProvides memory-friendly access to large collections of metadata.
-
index_class¶ alias of
StreamingIndex
-
papers¶
-
Module contents¶
The classes package provides the fundamental classes for working with
bibliographic data in Tethne.
paper |
A Paper represents a single bibliographic record. |
corpus |
A Corpus is a container for Papers. |
feature |
Classes in this module provide structures for additional data about Papers. |
graphcollection |
A GraphCollection is a set of graphs generated from a Corpus or model. |