tethne.networks package¶
Submodules¶
tethne.networks.authors module¶
Methods for generating networks in which authors are vertices.
author_papers |
A bi-partite graph containing Papers and their authors. |
coauthors |
A graph describing joint authorship in corpus. |
A bi-partite graph containing
Papers and their authors.
A graph describing joint authorship in
corpus.
tethne.networks.base module¶
-
tethne.networks.base.cooccurrence(corpus_or_featureset, featureset_name=None, min_weight=1, edge_attrs=['ayjid', 'date'], filter=None)[source]¶ A network of feature elements linked by their joint occurrence in papers.
tethne.networks.features module¶
Methods for building networks from terms in bibliographic records. This includes keywords, abstract terms, etc.
cooccurrence |
A network of feature elements linked by their joint occurrence in papers. |
mutual_information |
Generates a graph of features in featureset based on normalized pointwise mutual information (nPMI). |
keyword_cooccurrence |
|
topic_coupling |
-
tethne.networks.features.feature_cooccurrence(corpus, featureset_name, min_weight=1, filter=<function <lambda>>)[source]¶
-
tethne.networks.features.keyword_cooccurrence(corpus, min_weight=1, filter=<function <lambda>>)[source]¶
-
tethne.networks.features.mutual_information(corpus, featureset_name, min_weight=0.9, filter=<function <lambda>>)[source]¶ Generates a graph of features in
featuresetbased on normalized pointwise mutual information (nPMI).nPMI(i,j)=\frac{log(\frac{p_{ij}}{p_i*p_j})}{-1*log(p_{ij})}
...where p_i and p_j are the probabilities that features i and j will occur in a document (independently), and p_{ij} is the probability that those two features will occur in the same document.
tethne.networks.helpers module¶
Helper functions for generating networks.
citation_count |
Generates citation counts for all of the papers cited by papers. |
simplify_multigraph |
Simplifies a graph by condensing multiple edges between the same node pair into a single edge, with a weight attribute equal to the number of edges. |
top_cited |
Generates a list of the topn (or topn%) most cited papers. |
top_parents |
Returns a list of Paper that cite the topn most cited papers. |
-
tethne.networks.helpers.citation_count(papers, key='ayjid', verbose=False)[source]¶ Generates citation counts for all of the papers cited by papers.
Parameters: papers : list
A list of
Paperinstances.key : str
Property to use as node key. Default is ‘ayjid’ (recommended).
verbose : bool
If True, prints status messages.
Returns: counts : dict
Citation counts for all papers cited by papers.
-
tethne.networks.helpers.simplify_multigraph(multigraph, time=False)[source]¶ Simplifies a graph by condensing multiple edges between the same node pair into a single edge, with a weight attribute equal to the number of edges.
Parameters: graph : networkx.MultiGraph
E.g. a coauthorship graph.
time : bool
If True, will generate ‘start’ and ‘end’ attributes for each edge, corresponding to the earliest and latest ‘date’ values for that edge.
Returns: graph : networkx.Graph
A NetworkX
graph.
-
tethne.networks.helpers.top_cited(papers, topn=20, verbose=False)[source]¶ Generates a list of the topn (or topn%) most cited papers.
Parameters: papers : list
A list of
Paperinstances.topn : int or float {0.-1.}
Number (int) or percentage (float) of top-cited papers to return.
verbose : bool
If True, prints status messages.
Returns: top : list
A list of ‘ayjid’ keys for the topn most cited papers.
counts : dict
Citation counts for all papers cited by papers.
-
tethne.networks.helpers.top_parents(papers, topn=20, verbose=False)[source]¶ Returns a list of
Paperthat cite the topn most cited papers.Parameters: papers : list
A list of
Paperobjects.topn : int or float {0.-1.}
Number (int) or percentage (float) of top-cited papers.
verbose : bool
If True, prints status messages.
Returns: papers : list
A list of
Paperobjects.top : list
A list of ‘ayjid’ keys for the topn most cited papers.
counts : dict
Citation counts for all papers cited by papers.
tethne.networks.papers module¶
Methods for generating networks in which papers are vertices.
author_coupling |
|
bibliographic_coupling |
Generate a bibliographic coupling network. |
cocitation |
Generate a cocitation network. |
direct_citation |
A directed paper-citation network. |
topic_coupling |
-
tethne.networks.papers.bibliographic_coupling(corpus, min_weight=1, **kwargs)[source]¶ Generate a bibliographic coupling network.
Two papers are bibliographically coupled when they both cite the same, third, paper.
-
tethne.networks.papers.cocitation(corpus, min_weight=1, edge_attrs=['ayjid', 'date'], **kwargs)[source]¶ Generate a cocitation network.
A cocitation network is a network in which vertices are papers, and edges indicate that two papers were cited by the same third paper. CiteSpace is a popular desktop application for co-citation analysis, and you can read about the theory behind it here.
-
tethne.networks.papers.direct_citation(corpus, min_weight=1, **kwargs)[source]¶ A directed paper-citation network.
Direct-citation graphs are `directed acyclic graphs`__ in which vertices are papers, and each (directed) edge represents a citation of the target paper by the source paper. The
networks.papers.direct_citation()method generates both a global citation graph, which includes all cited and citing papers, and an internal citation graph that describes only citations among papers in the original dataset.
tethne.networks.topics module¶
Build networks from topics in a topic model.
The current implementation assumes that you are using a LDAModel.
-
tethne.networks.topics.cotopics(model, threshold=None, **kwargs)[source]¶ Two topics are coupled if they occur (above some
threshold) in the same document (s).Parameters: model :
LDAModelthreshold : float
Default:
2./model.Zkwargs : kwargs
Passed on to
cooccurrence().Returns:
-
tethne.networks.topics.distance(model, method='cosine', percentile=90, bidirectional=False, normalize=True, smooth=False, transform='log', **kwargs)[source]¶ Generate a network of
Papers based on a distance metric from scipy.spatial.distance using sparse-feature-vectors over the dimensions inmodel.The only two methods that will not work in this context are
hammingandjaccard.Distances are inverted to a similarity metric, which is log-transformed by default (see
transformparameter, below). Edges are included if they are at or above the ``percentile``th percentile.Parameters: model :
LDAModelorDTMModeldistance()usesmodel.itemandmodel.metadata.method : str
Name of a distance method from scipy.spatial.distance. See
analyze.features.distance()for a list of distance statistics.hammingorjaccardwill raise a RuntimeError.analyze.features.kl_divergence()is also available as ‘kl_divergence’.percentile : int
(default: 90) Edges are included if they are at or above the
percentilefor all distances in themodel.bidirectional : bool
(default: False) If True,
methodis calculated twice for each pair ofPapers ((i,j)and(j,i)), and the mean is used.normalize : bool
(default: True) If True, vectors over topics are normalized so that they sum to 1.0 for each
Paper.smooth : bool
(default: False) If True, vectors over topics are smoothed according to Bigi 2003. This may be useful if vectors over topics are very sparse.
transform : str
(default: ‘log’) Transformation to apply to similarity values before building the graph. So far only ‘log’ and None are supported.
Returns: Similarity values are included as edge weights. Node attributes are set using the fields in
model.metadata. Seenetworkx.Graph.__init__()Examples
>>> from tethne.networks import topics >>> thegraph = topics.distance(myLDAModel, 'cosine') >>> import tethne.writers as wr >>> wr.to_graphml(thegraph, '~./thegraph.graphml')
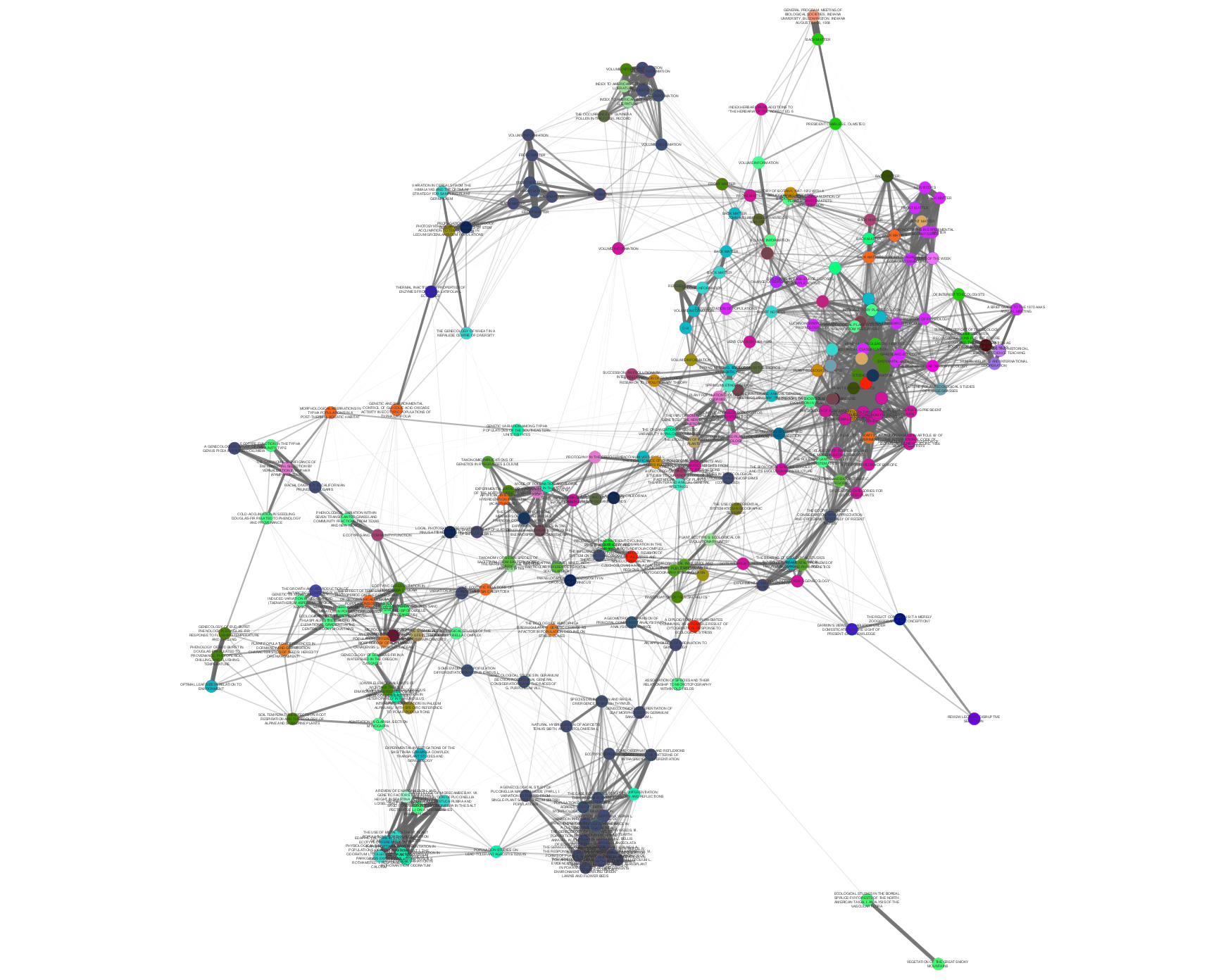
Edge weight and opacity indicate similarity. Node color indicates the journal in which each
Paperwas published. In this graph, papers published in the same journal tend to cluster together.
-
tethne.networks.topics.terms(model, threshold=0.01, **kwargs)[source]¶ Two terms are coupled if the posterior probability for both terms is greather than
thresholdfor the same topic.Parameters: model :
LDAModelthreshold : float
Default: 0.01
kwargs : kwargs
Passed on to
cooccurrence().Returns:
-
tethne.networks.topics.topic_coupling(model, threshold=None, **kwargs)[source]¶ Two papers are coupled if they both contain a shared topic above a
threshold.Parameters: model :
LDAModelthreshold : float
Default:
3./model.Zkwargs : kwargs
Passed on to
coupling().Returns:
Module contents¶
Methods for building networks from bibliographic data.
Each network relies on certain meta data in the Paper associated with
each document. Often we wish to construct a network with nodes representing
these documents and edges representing relationships between those documents,
but this is not always the case.
Where it is the case, it is recommended but not required that nodes are represented by an identifier from {ayjid, wosid, pmid, doi}. Each has certain benefits. If the documents to be networked come from a single database source such as the Web of Science, wosid is most appropriate. If not, using doi will result in a more accurate, but also more sparse network; while ayjid will result in a less accurate, but more complete network.
Any type of meta data from the Paper may be used as an identifier,
however.
We use “head” and “tail” nomenclature to refer to the members of a directed edge (x,y), x -> y, xy, etc. by calling x the “tail” and y the “head”.

