tethne.networks.features module¶
Methods for building networks from terms in bibliographic records. This includes keywords, abstract terms, etc.
| cooccurrence | Generates a cooccurrence graph for features in featureset. |
| mutual_information | Generates a graph of features in featureset based on normalized pointwise mutual information (nPMI). |
| keyword_cooccurrence | Generates a keyword cooccurrence network. |
| topic_coupling | Creates a network of words connected by implication in a common topic(s). |
- tethne.networks.features.cooccurrence(papers, featureset, filter=<function _filter at 0x108845c08>, graph=True, threshold=20, indexed_by='doi', **kwargs)[source]¶
Generates a cooccurrence graph for features in featureset.
filter is a method applied to each feature, used to determine whether a feature should be included in the graph before co-occurrence values are generated. This can cut down on computational expense. filter should accept the following parameters:
Parameter Description s Representation of the feature (e.g. a string). C The overall frequency of the feature in the Corpus. DC The number of documents in which the feature occurs. N Total number of documents in the Corpus. The default filter is:
>>> def _filter(s, C, DC, N): ... if C > 5 and DC > N*0.05 and len(s) > 4: ... return True ... return False
Parameters: papers : list
A list of Paper instances.
featurset : dict
A featureset from a Corpus.
filter : method
Method applied to each feature; should return True if the feature should be included, and False otherwise. See above.
graph : bool
(default: True) If False, returns a dictionary of co-occurrence values instead of a Graph.
threshold : int
(default: 20) Minimum co-occurrence value for inclusion in the Graph. If graph is False, this has no effect.
indexed_by : str
(default: ‘doi’) Field in Paper used as indexing values in featureset.
Returns: networkx.Graph or dict
See graph parameter, above.
- tethne.networks.features.keyword_cooccurrence(papers, threshold, connected=False, **kwargs)[source]¶
Generates a keyword cooccurrence network.
Parameters: papers : list
A list of Paper objects.
threshold : int
Minimum number of occurrences for a keyword pair to appear in graph.
connected : bool
If True, returns only the largest connected component.
Returns: k_coccurrence : networkx.Graph
A keyword coccurrence network.
Notes
Not thoroughly tested.
TODO
- Incorporate this into the featureset framework.
- tethne.networks.features.mutual_information(papers, featureset, filter=None, threshold=0.5, indexed_by='doi', **kwargs)[source]¶
Generates a graph of features in featureset based on normalized pointwise mutual information (nPMI).
nPMI(i,j)=\frac{log(\frac{p_{ij}}{p_i*p_j})}{-1*log(p_{ij})}...where p_i and p_j are the probabilities that features i and j will occur in a document (independently), and p_{ij} is the probability that those two features will occur in the same document.
Parameters: papers : list
A list of Paper instances.
featurset : dict
A featureset from a Corpus.
filter : method
Method applied to each feature prior to calculating co-occurrence. See cooccurrence().
threshold : float
(default: 0.5) Minimum nPMI for inclusion the graph.
indexed_by : str
(default: ‘doi’) Field in Paper used as indexing values in featureset.
Returns: graph : networkx.Graph
Examples
Using wordcount data from JSTOR Data-for-Research, we can generate a nPMI network as follows:
>>> from tethne.readers import dfr # Prep corpus. >>> MyCorpus = dfr.read_corpus(datapath+'/dfr', features=['uni']) >>> MyCorpus.filter_features('unigrams', 'u_filtered') >>> corpus.transform('u_filtered', 'u_tfidf') >>> from tethne.networks import features # Build graph. >>> graph = features.mutual_information(MyCorpus.all_papers(), 'u_tfidf') >>> from tethne.writers.graph import to_graphml # Export graph. >>> to_graphml(graph, '/path/to/my/graph.graphml')
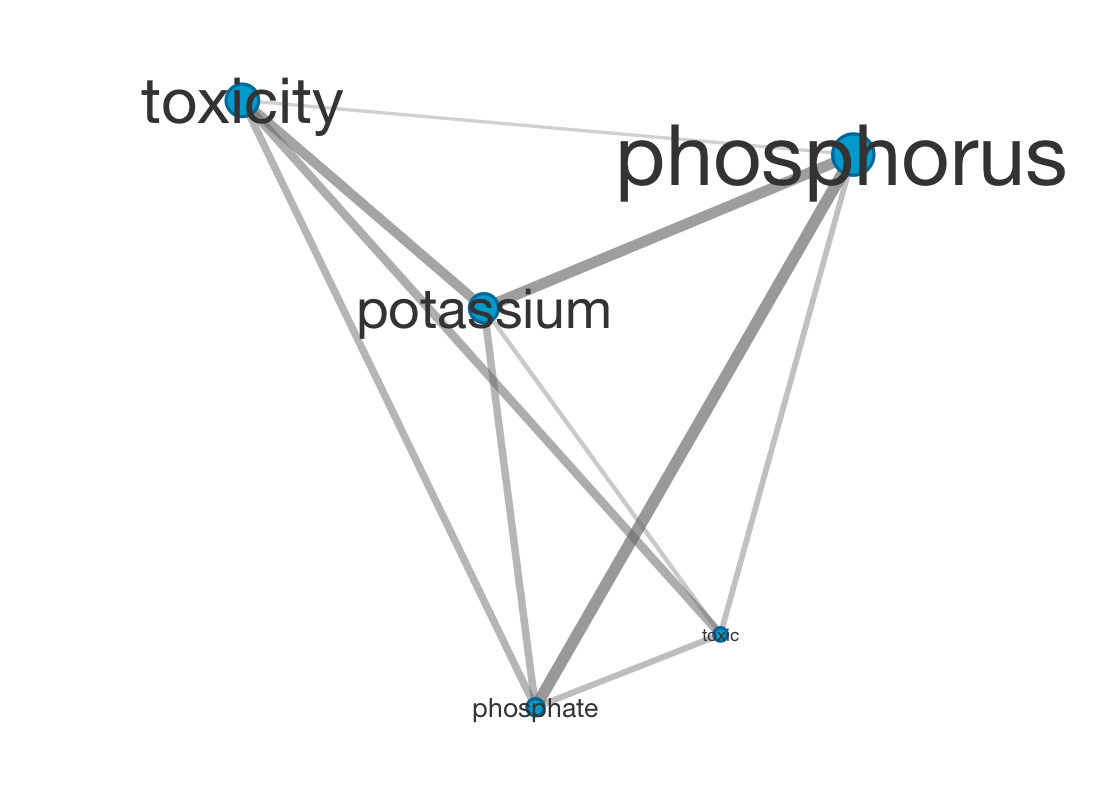
Here’s a small cluster from a similar graph, visualized in Cytoscape:
- tethne.networks.features.topic_coupling(model, threshold=0.005, **kwargs)[source]¶
Creates a network of words connected by implication in a common topic(s).
Parameters: model : LDAModel
threshold : float
Minimum P(W|T) for coupling.
Returns: tc : networkx.Graph
A topic-coupling graph, where nodes are terms.
Examples
>>> from tethne.networks import features >>> g = features.topic_coupling(MyLDAModel, threshold=0.015)
Here’s a similar network, visualized in Cytoscape:
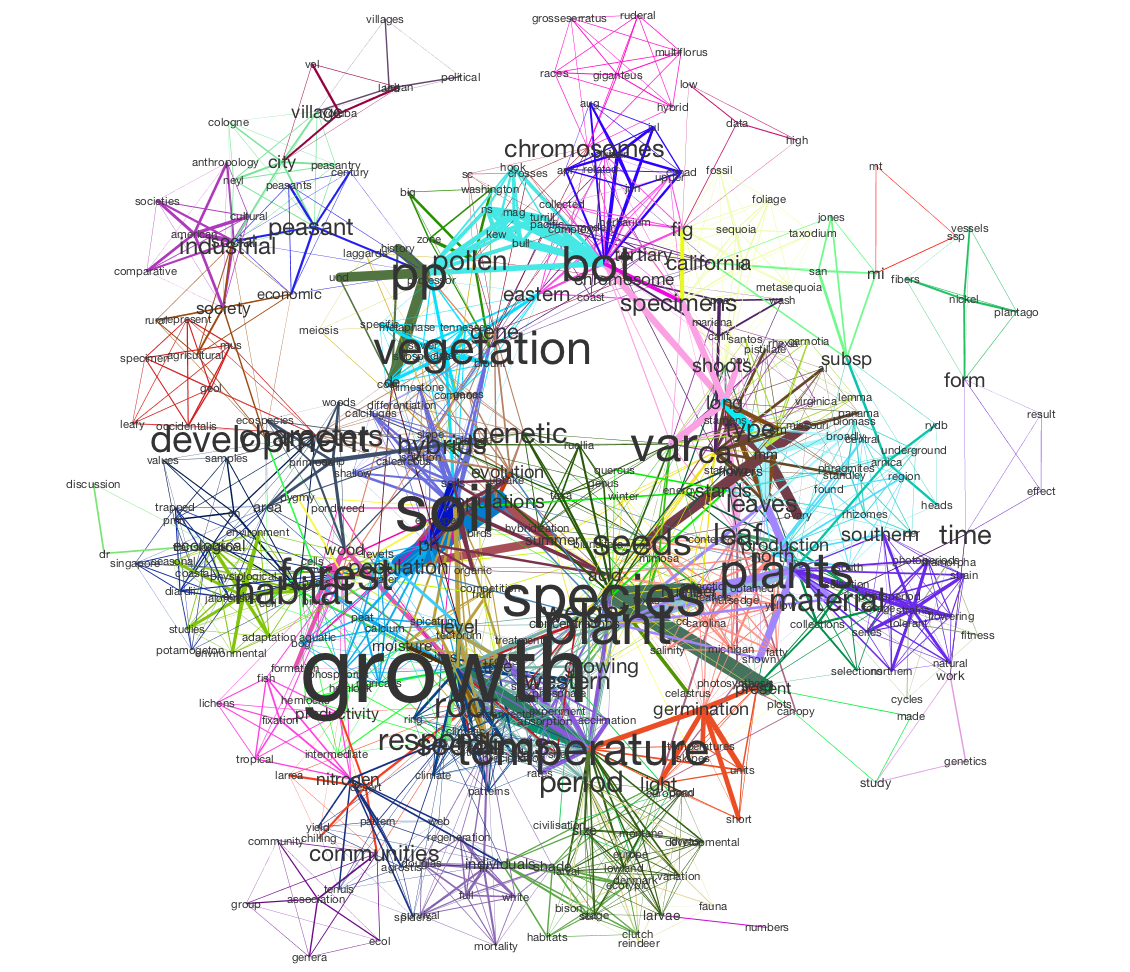
For details, see Generating and Visualizing Topic Models with Tethne and MALLET.